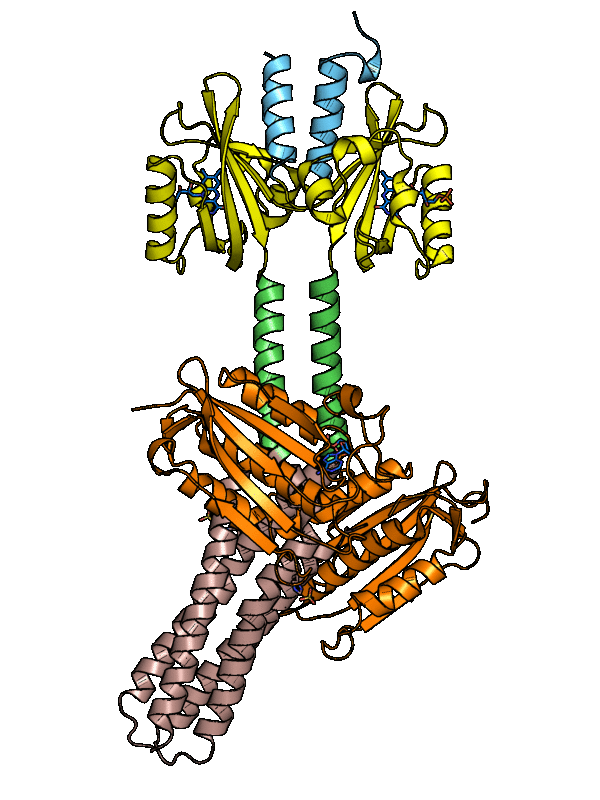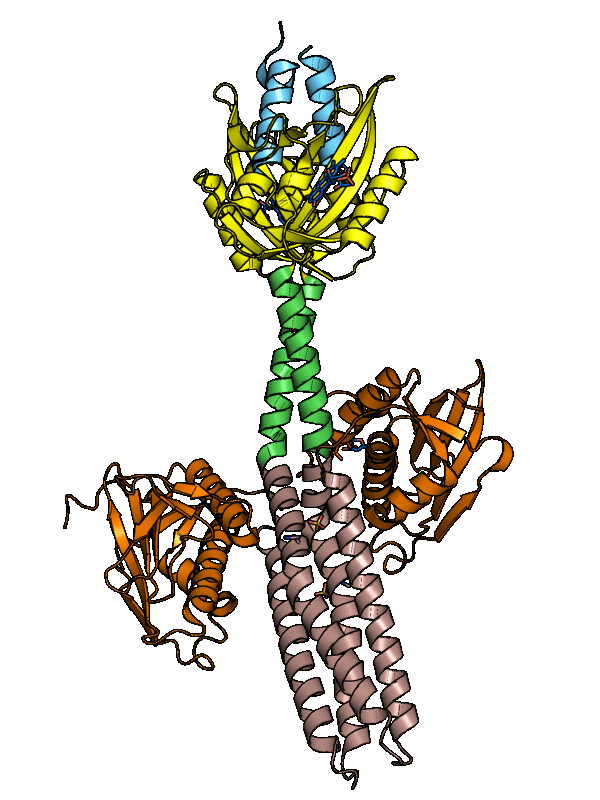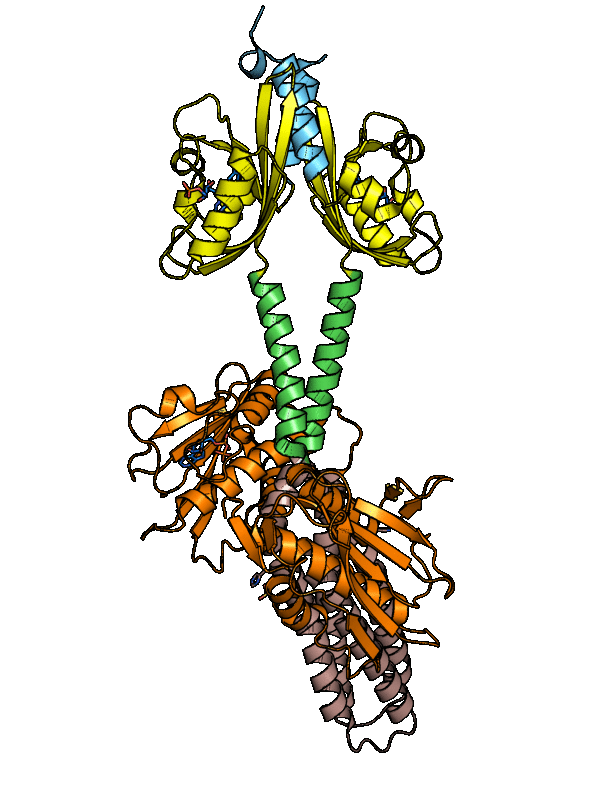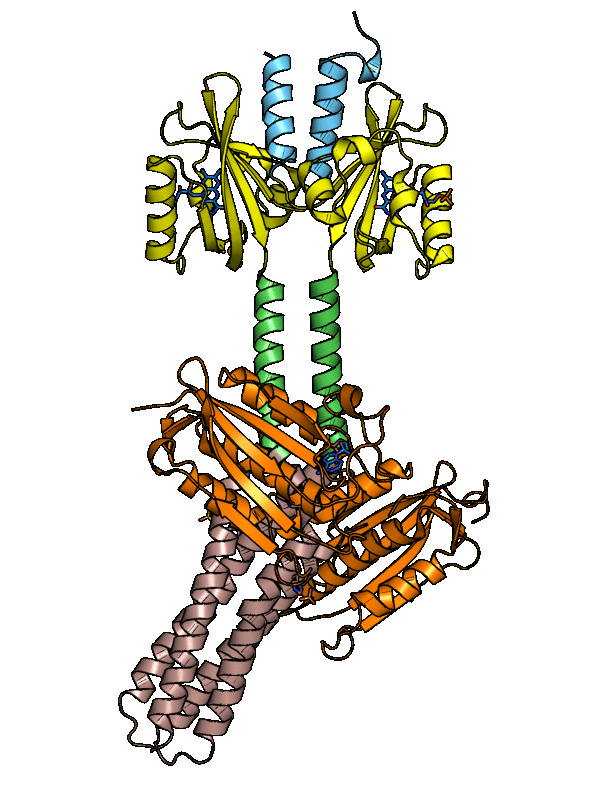
Structure of YF1. a Structure of YF1 in its dark-adapted state as

By A Mystery Man Writer
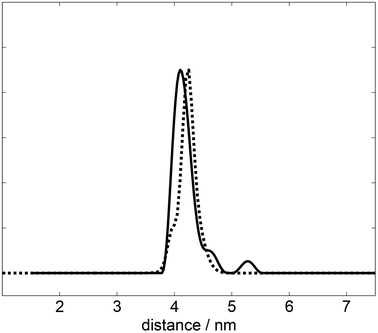
Download scientific diagram | Structure of YF1. a Structure of YF1 in its dark-adapted state as resolved by X-ray crystallography 13. The location of the different domains, of the flavin mononucleotide (FMN), of the cofactor adenosine diphosphate (ADP), and of the phosphoaccepting histidine 161 are indicated. b Light induced conformational changes of the LOV photosensor domain refined from X-ray solution scattering 22. The changes are maximal at the C-termini that feed into the Jα helices (dashed arrows). The coloring is according to the root mean square deviation of the alpha carbons from publication: Sequential conformational transitions and α-helical supercoiling regulate a sensor histidine kinase | Sensor histidine kinases are central to sensing in bacteria and in plants. They usually contain sensor, linker, and kinase modules and the structure of many of these components is known. However, it is unclear how the kinase module is structurally regulated. Here, we use | Secondary Protein Structure, Bacterial Proteins and Protein Conformation | ResearchGate, the professional network for scientists.
A structural model for the full-length blue light-sensing protein YtvA from Bacillus subtilis , based on EPR spectroscopy - Photochemical & Photobiological Sciences (RSC Publishing) DOI:10.1039/C3PP50128K

A) Structure of YF1 with a LOV photosensor dimer, consisting of A'α

Stephan NIEBLING, Postdoc, Applied Physics

Dimer asymmetry in the LOV-PAS and LOV-PAS-HK structures. (A) The
Research

Ralph DIENSTHUBER, Dr., Humboldt-Universität zu Berlin, Berlin, HU Berlin, Department of Biology

The six steps of the complete F1-ATPase rotary catalytic cycle
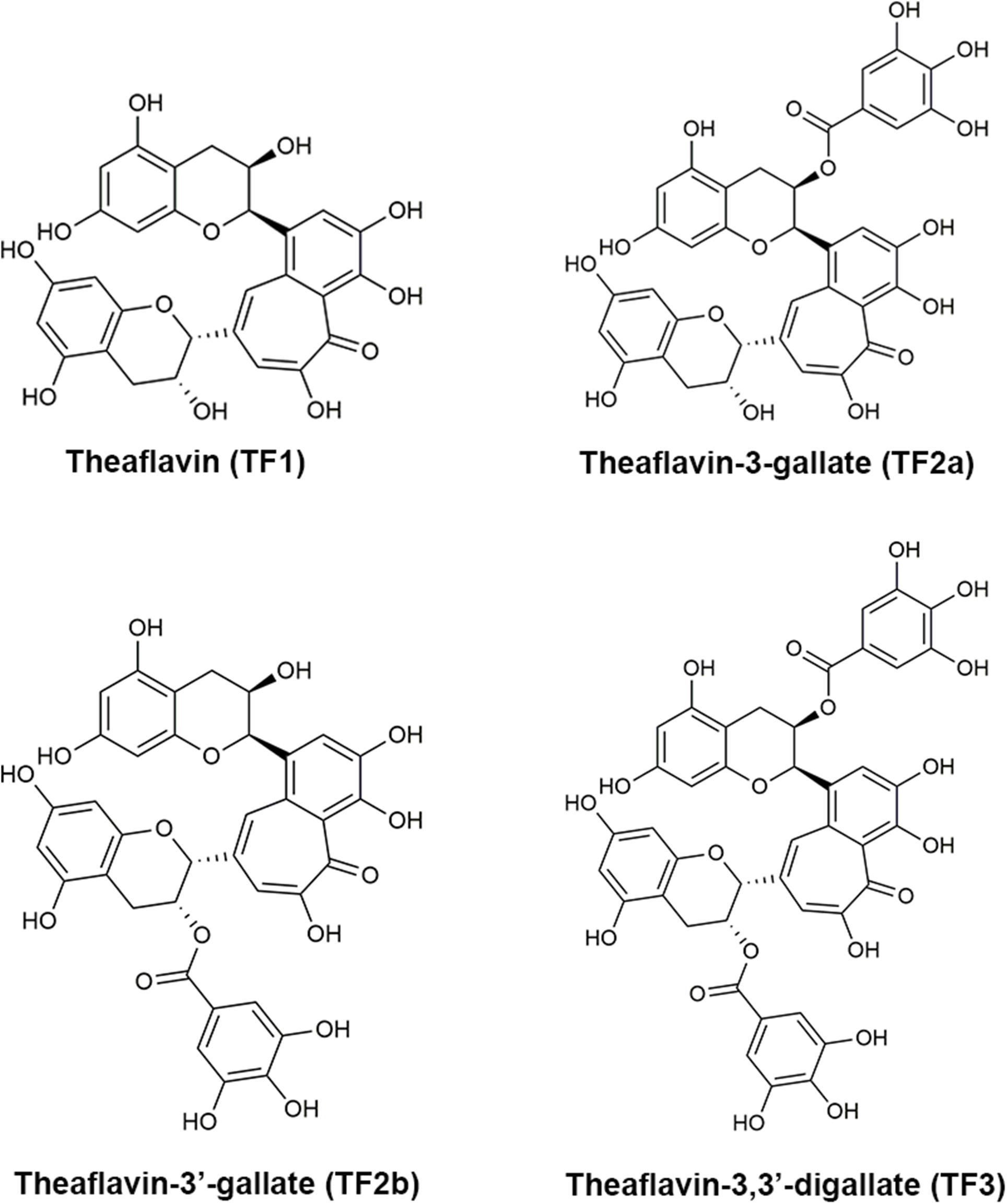
Frontiers Theaflavin-3,3′-Digallate Suppresses Biofilm Formation, Acid Production, and Acid Tolerance in Streptococcus mutans by Targeting Virulence Factors

Mode of autophosphorylation in bacteriophytochromes RpBphP2 and RpBphP3
- Reply to @maanyaownbusiness hope this helps! #fyp
- Lululemon Adapted State HR Jogger TF Zippered Pockets Size 10 Psychic 52655.

- Fat Adapted: What Does It Mean?

- Policy Brief – The Contribution of Marine Protected Areas to Climate Change Adaptation, State of the Evidence and Policy Recommendations - Ocean & Climate Platform
- Adapted state joggers in mineral blue (size 6) and oversized scuba 1/2 zip in heathered core ultra light grey (size M/L) : r/lululemon





/https%3A%2F%2Fd2umolmz9zcmr8.cloudfront.net%2Funsafe%2Fgenres%2Fbossa-nova_1.png)
