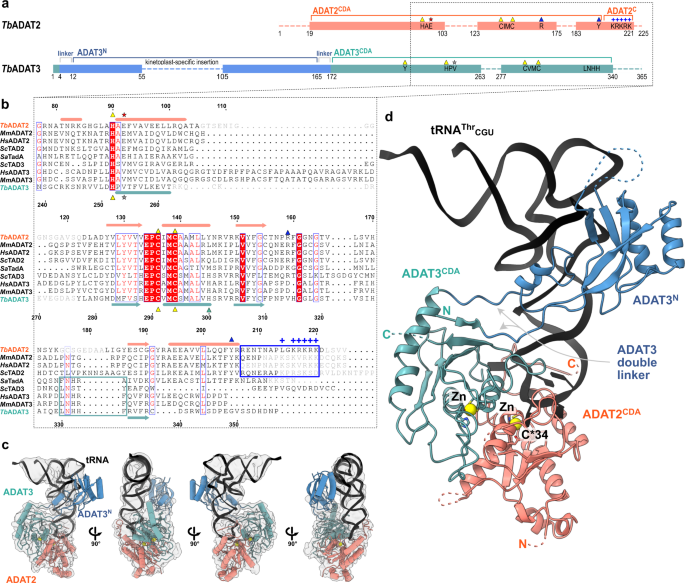
Structural basis for sequence-independent substrate selection by eukaryotic wobble base tRNA deaminase ADAT2/3
By A Mystery Man Writer

Distinct codon signature at the ribosome A-and P-sites in self

Structural basis for sequence-independent substrate selection by

Structural basis for anticodon recognition by discriminating

Turns in RNA motifs involving non-canonical base-pairs. (a) Views
Life, Free Full-Text

An ancient ancestral haplotype surrounding the ADAT3 locus. (A

Structural basis for sequence-independent substrate selection by

Sequence-independent substrate selection by the eukaryotic wobble

The possible single hydrogen-bonded arrangements of 32 Á 38 base

Modification of the adenosine residue into inosine at the wobble

A naturally occurring mini-alanyl-tRNA synthetase

Inosine induces stemness features in CAR-T cells and enhances

Structural basis of tRNA agmatinylation essential for AUA codon
RCSB PDB - 8AW3: Cryo-EM structure of the Tb ADAT2/3 deaminase in
- Relationship between gall diameter and the proportion of Eurosta

- Uses of National Household Travel Survey Data in - NHTS Home

- Level of importance of employment characteristics

- PDF) National survey of therapeutic orientation and associated factors of counselors and psychotherapists in China

- Remote Sensing, Free Full-Text
- Game Red Hoodie with Yayo White Classic Logo 3d Effect

- 6pcs Women Ladies Soft Comfortable Back Bra 2 Hooks / 3 Hooks / 4 Hooks Band Extension Strap Extender, White/Black/Khaki

- Garage Serena Seamless Deep Plunge Tank

- Elagalu Pine Cone Candle Stick - Rust (Set of 2) – Nkuku

- Isotoner Women's Brief Panties, 5-Pack
