K-mer analysis and genome size estimate
By A Mystery Man Writer

Difference in genome size estimation by genomescope depending on ploidy. · Issue #80 · KamilSJaron/smudgeplot · GitHub

A chromosome-level, haplotype-phased Vanilla planifolia genome highlights the challenge of partial endoreplication for accurate whole-genome assembly - ScienceDirect
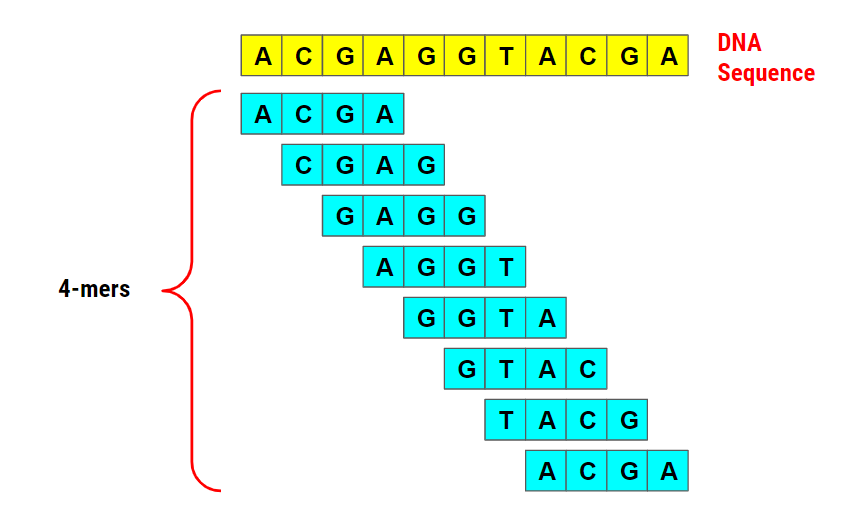
Bioinformatics 1: K-mer Counting. A challenging yet intriguing…, by Gunavaran Brihadiswaran, The Startup

K-Mer-Based Genome Size Estimation in Theory and Practice

Workflow (Illumina Inc), Bioz
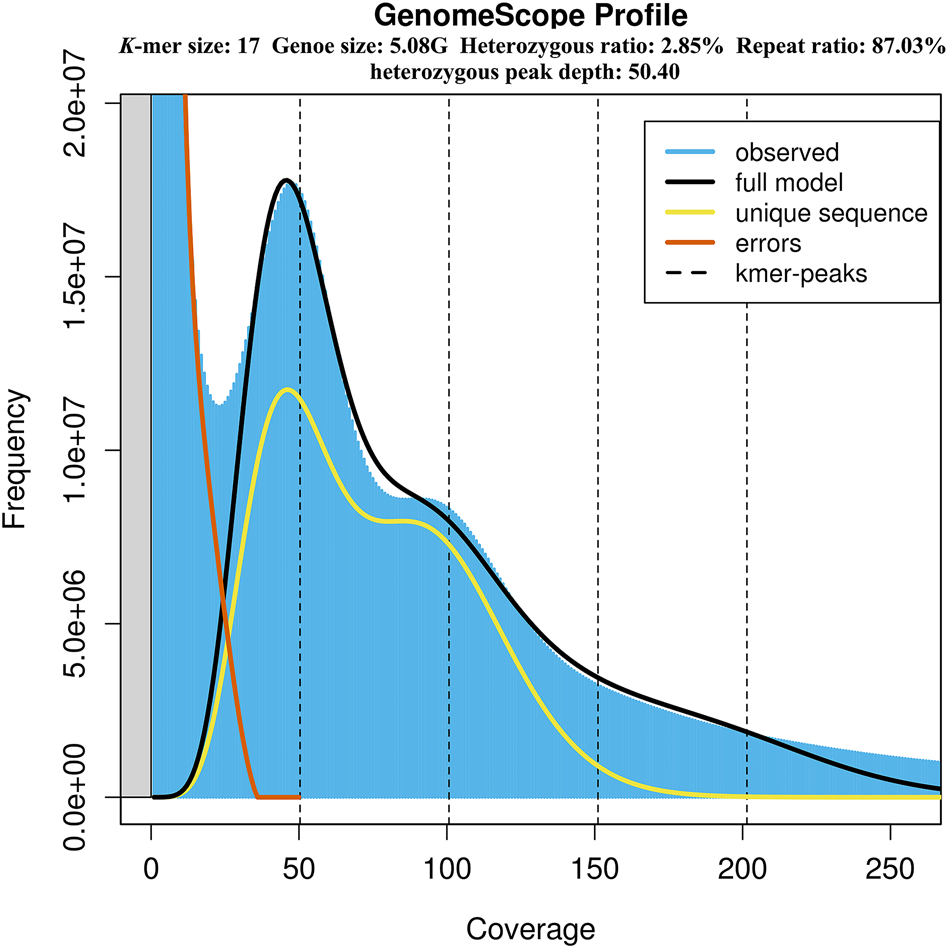
A first genome survey sequencing of alvinocaridid shrimp Shinkaicaris leurokolos in deep-sea hydrothermal vent environment, Journal of the Marine Biological Association of the United Kingdom

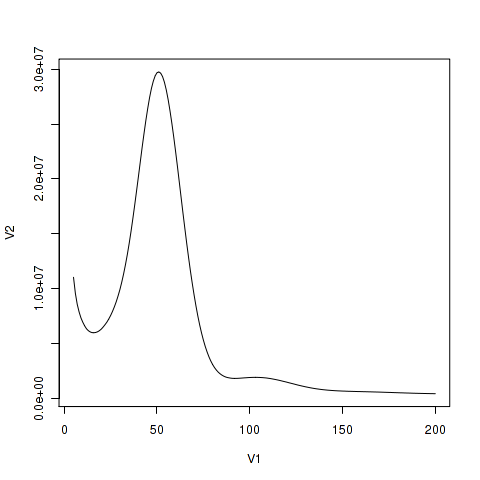
K-mer (K = 17) analysis for estimate genome size and heterozygote

GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes

iMOKA: k-mer based software to analyze large collections of sequencing data, Genome Biology

A beginner's guide to assembling a draft genome and analyzing structural variants with long-read sequencing technologies - ScienceDirect
k-mer - Wikipedia

K-Mer-Based Genome Size Estimation in Theory and Practice